CF Next Generation Sequencing
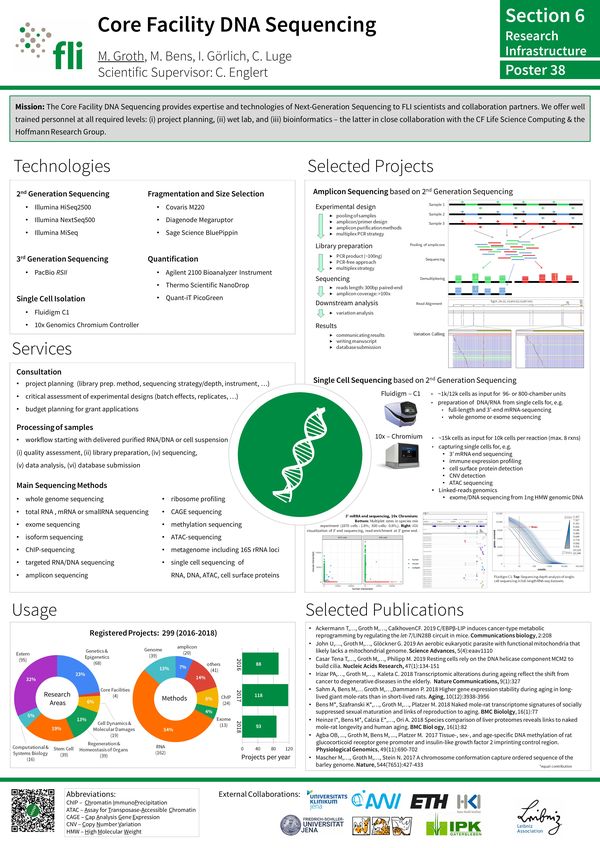
The Facility provides next-generation DNA sequencing (NGS) technology to the FLI. External users may also have access to the technology, e.g. upon collaborative arrangements. In addition to sample DNA/RNA quality check, library preparation and sequencing, the facility provides first-pass data processing, data quality check and analysis. A number of pipelines for deeper data analysis are established and available, also via the of the FLI.
Beyond analyzing whole genomes or subfractions thereof (e.g. exomes), NGS is ideally suited to provide insight into fundamental biological processes, such as transcription, translation, DNA methylation, histone modification, protein-nucleic acid interaction and much more.
The Facility operates the following technology platforms:
- Illumina NovaSeq6000
- Illumina NextSeq500
- Illumina MiSeq
- MinION (Oxford Nanopore)
- 10x Chromium X single-cell preparation
- Beckman-Cpoulter Biomek i7
- Covaris M220
- Diagenode Megaruptor
- Sage Science BluePippin
- Agilent 2100 Bioanalyzer
- Agilent 4200 Tapestation
- Thermo Scientific NanoDrop.

Full service:
- RNA-Seq (full-length transcripts): sequencing of polyA+/rRNA-RNA for analysis of differentially expressed genes or transcriptome assembly
- RNA-Seq (3'-ends): sequencing of 3'-ends of polyA+ RNA for analysis of differentially expressed genes or identification of transcription termination sites (TTS)
- CAGE-Seq: sequencing of 5'-ends of polyA+ RNA for identification of transcription start sites (TSS)
- SmallRNA-Seq: sequencing of small/micro RNAs for expression profiling
- ISO-Seq: sequencing of polyA+RNA for transcriptome assembly/transcript isoform discovery
- WES: whole exome sequencing for variant discovery
- WGS: whole genome sequencing for variant discovery or genome assembly
- WGBS/RRBS: whole genome/reduced representation bisulfite sequencing for DNA methylation analysis (5mC and/or 5hmC)
- 16s rDNA-Seq: sequencing of bacterial DNA regions coding for v3 and v4 of rRNA
Single-cell service:
- scRNA-Seq: sequencing of 3' ends of polyA+ RNAs of single cells for analysis of differentially expressed genes
- scATAC-Seq: genome-wide sequencing of transposase accessible DNA (open chromatin) in single-cell resolution
- scMultiome: combined scRNA-Seq and scATAC-Seq in single-nuclei resolution
Partial service:
- ChIP-Seq: sequencing of DNA following chromatin immunoprecipitation to identify protein binding sites
- ATAC-Seq: genome-wide sequencing of transposase accessible DNA (open chromatin)
- Amplicon-Seq: amplicon sequencing for variant discovery or DNA methylation analysis
- CUT&RUN / CUT&Taq: sequencing of DNA following antibody-targeted controlled cleavage by micrococcal nuclease/transposase
Internal users:
Resources, Cost Calculation, project sheets for requesting NGS services, quality check form for requesting QC of RNA/DNA/libraries
External users:
Please inquire.
Team*
* incomplete due to Data protection
For publications of individual CF members: publications search
Contact

Marco Groth
CF Manager
+49 3641 65-6054
cfngs@~@leibniz-fli.de
Christoph Englert
Scientific Supervisor
christoph.englert@~@leibniz-fli.de