Subarea 1: Stem Cell Aging
The individual research groups within Subarea 1 investigate the causes and consequences of stem cell aging. The research work spans from basic model organisms over genetic mouse models up to humanized mouse models engrafted with human stem cells.
According to the FLI, with the closure of two groups since 2016 the representation of invertebrate models of stem cell research was reduced in Subarea 1. The institute presumes that the recruitment of new groups should fill this gap.
The research is defined by four focus areas:
- Cell-intrinsic mechanisms limiting the function of aging stem and progenitor cells,
- Aging-associated alterations of stem cell niches and the systemic environment,
- Mechanisms of clonal selection and epigenetic drifts in stem cell aging, and
- Microbiota- and metabolism-induced impairments in stem cell function during aging (in context of the new focus area Microbiota and Aging currently being built up within Subarea 2).
Research focus of Subarea 1.
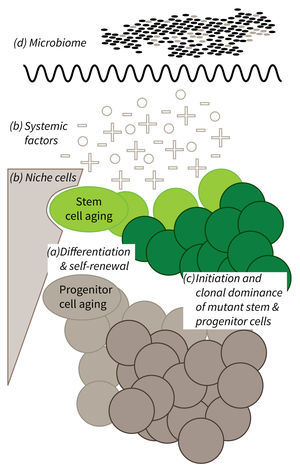
a) It is currently not well understood what mechanisms impair cellular functions in aging. b) The relative contribution of niche cells and systemic acting factors on stem cell aging have yet to be determined in different tissues. c) Clonal expansion of mutant cells associates with disease development in aging humans. Mechanistically, the process remains poorly understood. Changes in color intensity depict clonal dominance originating from stem (green) or progenitor cells (gray). d) Emerging evidences indicate that aging associated alter ations in microbiota influence stem cell function and vice versa.
Publications
(since 2016)
2018
- A Boolean network of the crosstalk between IGF and Wnt signaling in aging satellite cells.
Siegle* L, Schwab* JD, Kühlwein* SD, Lausser L, Tümpel S, Pfister AS, Kühl** M, Kestler** HA
PLoS One 2018, 13(3), e0195126 * equal contribution, ** co-senior authors - Senescence mirrors the extent of liver fibrosis in chronic hepatitis C virus infection.
Wandrer F, Han B, Liebig S, Schlue J, Manns MP, Schulze-Osthoff K, Bantel H
Aliment Pharmacol Ther 2018, 48(3), 270-80
2017
- Identification of novel Hox target genes with essential roles in hematopoietic stem cell self-renewal and differentiation
Baig AH
Dissertation 2017, Jena, Germany - RSK-mediated nuclear accumulation of the cold-shock Y-box protein-1 controls proliferation of T cells and T-ALL blasts.
Gieseler-Halbach S, Meltendorf S, Pierau M, Weinert S, Heidel FH, Fischer T, Handschuh J, Braun-Dullaeus RC, Schrappe M, Lindquist JA, Mertens PR, Thomas U, Brunner-Weinzierl MC
Cell Death Differ 2017, 24(2), 371-83 - Planarian finds time(less) to fight infection.
Gutiérrez-Gutiérrez Ó, Felix DA, González-Estévez C
Virulence 2017, 8(7), 1043-8 - mTORC1 Activation during Repeated Regeneration Impairs Somatic Stem Cell Maintenance.
Haller S, Kapuria S, Riley RR, O'Leary MN, Schreiber KH, Andersen JK, Melov S, Que J, Rando TA, Rock J, Kennedy BK, Rodgers JT, Jasper H
Cell Stem Cell 2017, 21(6), 806-818.e5 - Gene dosage reductions of Trf1 and/or Tin2 induce telomere DNA damage and lymphoma formation in aging mice.
Hartmann K, Illing A, Leithäuser F, Baisantry A, Quintanilla-Martinez L, Rudolph KL
Leukemia 2017, 31(12), 2853 Erratum for Leukemia 2016 volume 30 page 749 - Protein phosphatase 4 regulatory subunit 2 (PPP4R2) is recurrently deleted in acute myeloid leukemia and required for efficient DNA double strand break repair.
Herzig JK, Bullinger L, Tasdogan A, Zimmermann P, Schlegel M, Teleanu V, Weber D, Rücker FG, Paschka P, Dolnik A, Schneider E, Kuchenbauer F, Heidel FH, Buske C, Döhner H, Döhner K, Gaidzik VI
Oncotarget 2017, 8(56), 95038-53 - Hacking the stem cell niche.
Lane SW, Heidel FH
Blood 2017, 129(22), 2951-2 - Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion.
Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, Wang ZQ, Kestler HA, Fagagna Fdd, Günes C, Rudolph KL
EMBO J 2017, 36(19), 2922-4 Erratum for EMBO J 2015 volume 34 page 1371