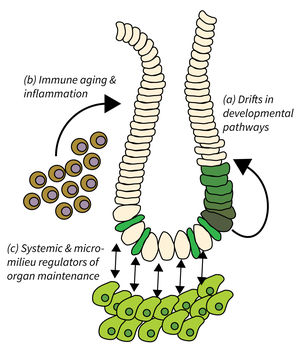
Subarea 2: Regeneration and Homeostasis of Organs in Aging
The main goal of Subarea 2 is to identify cellular and molecular pathways used to ensure effective organ maintenance and repair, and to unravel the mechanisms of their deterioration during aging. While stem cells are important for organ homeostasis, this Subarea does not per se directly addresses stem cell aging but rather focusses on the following focus areas:
- Drifts in developmental pathways limiting organ maintenance in aging,
- Immune aging and inflammation, and
- Systemic and micro-milieu regulators of organ maintenance, regeneration, and disease development.
Research focus of Subarea 2
Organ maintenance is regulated by local and systemic factors, which are subject to aging-associated changes. Research of Subarea 2 focuses on the following research areas: a) Genetic and epigenetic modulation of developmental pathways has been shown to contribute to progressive aging and disease. It is critical to delineate mechanisms and consequences of aging-associated drifts to better understand organ maintenance during aging. b) Immunoaging and chronic inflammation elicits negative effects through reduced immune surveillance and aberrant organ repair and maintenance; all of which contributes to the evolution of organ pathologies and diseases during organismal aging. c) Furthermore, aging-associated alterations in systemic and extracellular factors derived from metabolic changes, microbiota alterations, chronic inflammation, senescent, or damaged cells might impinge on disease development and tumor initiation.
Publications
(since 2016)
2018
- Thyroid Hormone Transporters MCT8 and OATP1C1 Control Skeletal Muscle Regeneration.
Mayerl S, Schmidt M, Doycheva D, Darras VM, Hüttner SS, Boelen A, Visser TJ, Kaether C, Heuer** H, von Maltzahn** J
Stem Cell Reports 2018, 10(6), 1959-74 ** co-corresponding authors - Reduced expression of C/EBPβ-LIP extends health- and lifespan in mice.
Müller* C, Zidek* LM, Ackermann T, de Jong T, Liu P, Kliche V, Zaini MA, Kortman G, Harkema L, Verbeek DS, Tuckermann JP, von Maltzahn J, de Bruin A, Guryev V, Wang ZQ, Calkhoven CF
Elife 2018, 7, e34985 * equal contribution - Survivin antagonizes chemotherapy-induced cell death of colorectal cancer cells.
Rauch A, Carlstedt A, Emmerich C, Mustafa AHM, Göder A, Knauer SK, Linnebacher M, Heinzel T, Krämer OH
Oncotarget 2018, 9(45), 27835-50 - The actin binding protein Ezrin controls astrocyte functions in the healthy and injured brain.
Schacke S
Dissertation 2018, Jena, Germany - Gamma secretase dependent release of the CD44 cytoplasmic tail upregulates IFI16 in cd44-/- tumor cells, MEFs and macrophages.
Schultz* K, Grieger (Lindner)* C, Li Y, Urbánek P, Ruschel A, Minnich K, Bruder D, Gereke M, Sechi A, Herrlich P
PLoS One 2018, 13(12), e0207358 * equal contribution - Neuropathies in the setting of Neurofibromatosis tumor syndromes: Complexities and opportunities.
Schulz A, Grafe P, Hagel C, Bäumer P, Morrison H, Mautner VF, Farschtschi S
Exp Neurol 2018, 299(Pt B), 334-44 - Traditional and systems biology based drug discovery for the rare tumor syndrome neurofibromatosis type 2.
Synodos for NF2 Consortium, Allaway R, Angus SP, Beauchamp RL, Blakeley JO, Bott M, Burns SS, Carlstedt A, Chang LS, Chen X, Clapp DW, Desouza PA, Erdin S, Fernandez-Valle C, Guinney J, Gusella JF, Haggarty SJ, Johnson GL, La Rosa S, Morrison H, Petrilli AM, Plotkin SR, Pratap A, Ramesh V, Sciaky N, Stemmer-Rachamimov A, Stuhlmiller TJ, Talkowski ME, Welling DB, Yates CW, Zawistowski JS, Zhao WN
PLoS One 2018, 13(6), e0197350 - Neurofibromatosis type 2 tumor suppressor protein is expressed in oligodendrocytes and regulates cell proliferation and process formation.
Toledo* A, Grieger* E, Karram K, Morrison H, Baader SL
PLoS One 2018, 13(5), e0196726 * equal contribution - Induction chemoradiotherapy versus induction chemotherapy for potentially resectable stage IIIA (N2) non-small cell lung cancer: a systematic review and meta-analysis.
Tong S, Qin Z, Wan M, Zhang L, Cui Y, Yao Y
J Thorac Dis 2018, 10(4), 2428-36 - A p300 and SIRT1 Regulated Acetylation Switch of C/EBPα Controls Mitochondrial Function.
Zaini MA, Müller C, de Jong TV, Ackermann T, Hartleben G, Kortman G, Gührs KH, Fusetti F, Krämer OH, Guryev V, Calkhoven CF
Cell Rep 2018, 22(2), 497-511