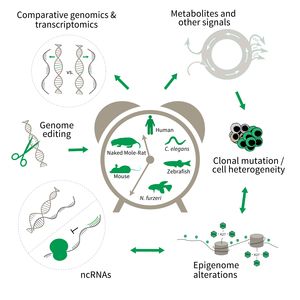
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influ ence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2022
- Androgen-Induced MIG6 Regulates Phosphorylation of Retinoblastoma Protein and AKT to Counteract Non-Genomic AR Signaling in Prostate Cancer Cells.
Schomann T, Mirzakhani K, Kallenbach J, Lu J, Rasa SMM, Neri F, Baniahmad A
Biomolecules 2022, 12(8), 1048 - Moderation of rRNA gene activity triggers metabolic adaptation promoting geroprotection in Caenorhabditis elegans
Sharifi MS
Dissertation 2022, Jena, Germany - Atlas of the aging mouse large intestine
Sirvinskas D
Dissertation 2022, Jena, Germany - Single-cell atlas of the aging mouse colon.
Širvinskas D, Omrani O, Lu J, Rasa M, Krepelova A, Adam L, Kaeppel S, Sommer F, Neri F
iScience 2022, 25(5), 104202 - A genome-wide CRISPR activation screen reveals Hexokinase 1 as a critical factor in promoting resistance to multi-kinase inhibitors in hepatocellular carcinoma cells.
Sofer S, Lamkiewicz K, Armoza Eilat S, Partouche S, Marz M, Moskovits N, Stemmer SM, Shlomai A, Sklan EH
FASEB J 2022, 36(3), e22191 - Sex chromosome differentiation via changes in the Y chromosome repeat landscape in African annual killifishes Nothobranchius furzeri and N. kadleci.
Štundlová J, Hospodářská M, Lukšíková K, Voleníková A, Pavlica T, Altmanová M, Richter A, Reichard M, Dalíková M, Pelikánová Š, Marta A, Simanovsky SA, Hiřman M, Jankásek M, Dvořák T, Bohlen J, Ráb P, Englert C, Nguyen P, Sember A
Chromosome Res 2022, 30(4), 309-33 - The Mating Pattern of Captive Naked Mole-Rats Is Best Described by a Monogamy Model
Szafranski* K, Wetzel* M, Holtze S, Büntjen I, Lieckfeldt D, Ludwig A, Huse K, Platzer M, Hildebrandt T
FRONT ECOL EVOL 2022, 10, 855688 * equal contribution - Characterization of RNA content in individual phase-separated coacervate microdroplets.
Wollny D, Vernot B, Wang J, Hondele M, Safrastyan A, Aron F, Micheel J, He Z, Hyman A, Weis K, Camp JG, Tang TYD, Treutlein B
Nat Commun 2022, 13(1), 2626 - The Role of Non-Coding RNAs in the Human Placenta.
Žarković* M, Hufsky* F, Markert** UR, Marz** M
Cells 2022, 11(9), 1588 * equal contribution, ** co-senior authors