Subarea 3: Genetics and Epigenetics of Aging
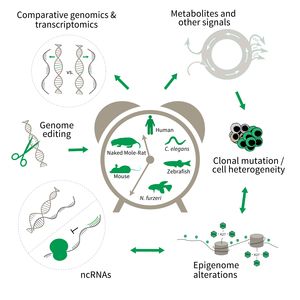
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influ ence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2024
- Detection of reproducible liver cancer specific ligand-receptor signaling in blood.
Safrastyan* ** A, Wollny* ** D
Front Bioinform 2024, 4, 1332782 * equal contribution, ** co-senior authors - Autoantigen-specific CD4+ T cells acquire an exhausted phenotype and persist in human antigen-specific autoimmune diseases.
Saggau C, Bacher P, Esser D, Rasa M, Meise S, Mohr N, Kohlstedt N, Hutloff A, Schacht SS, Dargvainiene J, Martini GR, Stürner KH, Schröder I, Markewitz R, Hartl J, Hastermann M, Duchow A, Schindler P, Becker M, Bautista C, Gottfreund J, Walter J, Polansky JK, Yang M, Naghavian R, Wendorff M, Schuster EM, Dahl A, Petzold A, Reinhardt S, Franke A, Wieczorek M, Henschel L, Berger D, Heine G, Holtsche M, Häußler V, Peters C, Schmidt E, Fillatreau S, Busch DH, Wandinger KP, Schober K, Martin R, Paul F, Leypoldt F, Scheffold A
Immunity 2024 (epub ahead of print) - The Greenland shark (Somniosus microcephalus) genome provides insights into extreme longevity
Sahm A, Cherkasov* A, Liu* H, Voronov D, Siniuk K, Schwarz R, Ohlenschlaeger O, Foerste S, Bens M, Groth M, Goerlich I, Paturej S, Klages S, Braendl B, Olsen J, Bushnell P, Bech Poulsen A, Ferrando S, Garibaldi F, Lorenzo Drago D, Terzibasi Tozzini E, Mueller FJ, Fischer M, Kretzmer H, Domenici** P, Fleng Steffensen** J, Cellerino** A, Hoffmann** S
bioRxiv 2024, 10.1101/2024.09.09.611499 * equal contribution, ** co-corresponding authors - Hydra has mammal-like mutation rates facilitating fast adaptation despite its nonaging phenotype.
Sahm A, Riege K, Groth M, Bens M, Kraus J, Fischer M, Kestler H, Englert C, Schaible R, Platzer M, Hoffmann S
Genome Res 2024, 34(12), 2217-28 - Reducing the metabolic burden of rRNA synthesis promotes healthy longevity in Caenorhabditis elegans.
Sharifi* S, Chaudhari* P, Martirosyan A, Eberhardt AO, Witt F, Gollowitzer A, Lange L, Woitzat Y, Okoli EM, Li H, Rahnis N, Kirkpatrick J, Werz O, Ori A, Koeberle A, Bierhoff** H, Ermolaeva** M
Nat Commun 2024, 15(1), 1702 * equal contribution, ** co-corresponding authors - Tau mediates the reshaping of the transcriptional landscape toward intermediate Alzheimer's disease stages.
Siano G, Varisco M, Terrigno M, Wang C, Scarlatti A, Iannone V, Groth M, Galas MC, Hoozemans JJM, Cellerino A, Cattaneo A, Di Primio C
Front Cell Dev Biol 2024, 12, 1459573 - Comprehensive survey of conserved RNA secondary structures in full-genome alignment of Hepatitis C virus.
Triebel S, Lamkiewicz K, Ontiveros N, Sweeney B, Stadler PF, Petrov AI, Niepmann M, Marz M
Sci Rep 2024, 14(1), 15145 - Bidirectional Impact of miR-29a modulation on Memory Stability in the Adult Brain
Viglione A, Giannuzzi C, Putignano E, Mazziotti R, Cellerino A, Pizzorusso T
bioRxiv 2024, https://doi.org/10.1101/2024.11. - Maternal perceived stress and green spaces during pregnancy are associated with adult offspring gene (NR3C1 and IGF2/H19) methylation patterns in adulthood: A pilot study.
Vos S, Van den Bergh BRH, Martens DS, Bijnens E, Shkedy Z, Kindermans H, Platzer M, Schwab M, Nawrot TS
Psychoneuroendocrinology 2024, 167, 107088 - UFMylation of CDK5RAP2/MCPH3 regulates mitosis
Wu X
Dissertation 2024, Jena, Germany