Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
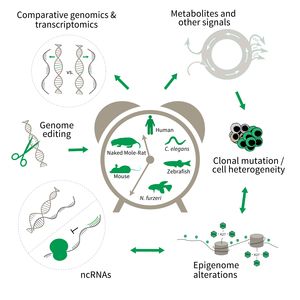
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influ ence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2016
- From the bush to the bench: the annual Nothobranchius fishes as a new model system in biology.
Cellerino A, Valenzano DR, Reichard M
Biol Rev Camb Philos Soc 2016, 91(2), 511-33 - Neurotrophin-4 in the brain of adult Nothobranchius furzeri.
D'Angelo L, Avallone L, Cellerino A, de Girolamo P, Paolucci M, Varricchio E, Lucini C
Ann Anat 2016, 207, 47-54 - Discovering miRNA regulatory networks in Holt-Oram Syndrome using a Zebrafish model
D'Aurizio R, Russo F, Chiavacci E, Baumgart M, Groth M, D'Onofrio M, Arisi I, Rainaldi G, Pitto L, Pellegrini M
Front. Bioeng. Biotechnol. 2016, 4, Article 60 - Analysis of cultivable microbiota and diet intake pattern of the long-lived naked mole-rat.
Debebe T, Holtze S, Morhart M, Hildebrandt TB, Rodewald S, Huse K, Platzer M, Wyohannes D, Yirga S, Lemma A, Thieme R, König B, Birkenmeier G
Gut Pathog 2016, 8, 25 - Ethyl Pyruvate: An Anti-Microbial Agent that Selectively Targets Pathobionts and Biofilms.
Debebe T, Krüger M, Huse K, Kacza J, Mühlberg K, König B, Birkenmeier G
PLoS One 2016, 11(9), e0162919 - Low sulfide levels and a high degree of cystathionine β-synthase (CBS) activation by S-adenosylmethionine (SAM) in the long-lived naked mole-rat.
Dziegelewska M, Holtze S, Vole C, Wachter U, Menzel U, Morhart M, Groth M, Szafranski K, Sahm A, Sponholz C, Dammann P, Huse K, Hildebrandt** T, Platzer** M
Redox Biol 2016, 8, 192-8 ** co-senior authors - Prediction of conserved long-range RNA-RNA interactions in full viral genomes.
Fricke M, Marz M
Bioinformatics 2016, 32(19), 2928-35 - High Copy Numbers of Beta-Defensin Cluster on 8p23.1 Confer Genetic Susceptibility and Modulate the Physical Course of Hidradenitis Suppurativa/Acne Inversa.
Giamarellos-Bourboulis* EJ, Platzer* M, Karagiannidis* I, Kanni T, Nikolakis G, Ulrich J, Bellutti M, Gollnick H, Bauer M, Zouboulis CC, Huse K
J Invest Dermatol 2016, 136(8), 1592-8 * equal contribution - Genomic engineering of Wilms tumour genes in Danio rerio
Große A
Dissertation 2016, Jena, Germany - Age-Dependent Changes of Monocarboxylate Transporter 8 Availability in the Postnatal Murine Retina.
Henning Y, Szafranski K
Front Cell Neurosci 2016, 10, 205