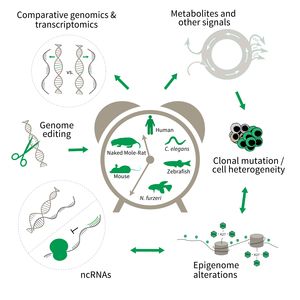
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2020
- Wt1 Positive dB4 Neurons in the Hindbrain Are Crucial for Respiration
Schnerwitzki* D, Hayn* C, Perner B, Englert C
Front. Neurosci. 2020, 14, 529487 * equal contribution - The circuitry between ribosome biogenesis and translation in stem cell function and ageing.
Sharifi'* S, da Costa* HFR, Bierhoff H
Mech Ageing Dev 2020, 189, 111282 * equal contribution - Intestinal stem cells heterogeneity and clonal dominance during aging: two faces of the same coin?
Sirvinskas* D, Annunziata* F, Neri F
Mech Ageing Dev 2020, 189, 111247 * equal contribution - Tissue-specific Gene Expression Changes Are Associated with Aging in Mice.
Srivastava* A, Barth* E, Ermolaeva MA, Guenther M, Frahm C, Marz** M, Witte** OW
Genomics Proteomics Bioinformatics 2020, 18(4), 430-42 * equal contribution, ** co-corresponding authors - Nothobranchius annual killifishes
Terzibasi Tozzini E, Cellerino A
Evodevo 2020, 11(1), 25 - Comparison of Multiscale Imaging Methods for Brain Research.
Tröger J, Hoischen C, Perner B, Monajembashi S, Barbotin A, Löschberger A, Eggeling C, Kessels** MM, Qualmann** B, Hemmerich** P
Cells 2020, 9(6), E1377 ** co-corresponding authors - Active neutrophil responses counteract Candida albicans burn wound infection of ex vivo human skin explants.
von Müller C, Bulman F, Wagner L, Rosenberger D, Marolda A, Kurzai O, Eißmann P, Jacobsen ID, Perner B, Hemmerich P, Vylkova S
Sci Rep 2020, 10(1), 21818 - Expansion of the renal capsular stroma, ureteric bud branching defects and cryptorchidism in mice with Wilms tumor 1 gene deletion in the stromal compartment of the developing kidney.
Weiss AC, Rivera-Reyes R, Englert C, Kispert A
J Pathol 2020, 252(3), 290-303
2019
- C/EBPβ-LIP induces cancer-type metabolic reprogramming by regulating the let-7 /LIN28B circuit in mice.
Ackermann T, Hartleben* G, Müller* C, Mastrobuoni G, Groth M, Sterken BA, Zaini MA, Youssef SA, Zuidhof HR, Krauss SR, Kortman G, de Haan G, de Bruin A, Wang ZQ, Platzer M, Kempa S, Calkhoven CF
Commun Biol 2019, 2, 208 * equal contribution - Conserved aging-related signatures of senescence and inflammation in different tissues and species.
Barth* E, Srivastava* A, Stojiljkovic* M, Frahm C, Axer H, Witte** OW, Marz** M
Aging (Albany NY) 2019, 11(19), 8556-72 * equal contribution, ** co-senior authors