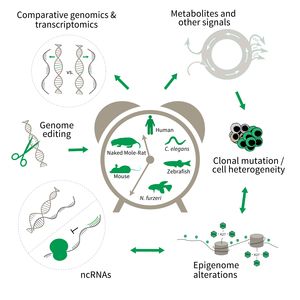
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2017
- A chromosome conformation capture ordered sequence of the barley genome.
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, Bayer M, Ramsay L, Liu H, Haberer G, Zhang XQ, Zhang Q, Barrero RA, Li L, Taudien S, Groth M, Felder M, Hastie A, Šimková H, Staňková H, Vrána J, Chan S, Muñoz-Amatriaín M, Ounit R, Wanamaker S, Bolser D, Colmsee C, Schmutzer T, Aliyeva-Schnorr L, Grasso S, Tanskanen J, Chailyan A, Sampath D, Heavens D, Clissold L, Cao S, Chapman B, Dai F, Han Y, Li H, Li X, Lin C, McCooke JK, Tan C, Wang P, Wang S, Yin S, Zhou G, Poland JA, Bellgard MI, Borisjuk L, Houben A, Doležel J, Ayling S, Lonardi S, Kersey P, Langridge P, Muehlbauer GJ, Clark MD, Caccamo M, Schulman AH, Mayer KFX, Platzer M, Close TJ, Scholz U, Hansson M, Zhang G, Braumann I, Spannagl M, Li C, Waugh R, Stein N
Nature 2017, 544(7651), 427-33 - The Wilms tumor protein Wt1 contributes to female fertility by regulating oviductal proteostasis.
Nathan A, Reinhardt P, Kruspe D, Jörß T, Groth M, Nolte H, Habenicht A, Herrmann J, Holschbach V, Toth B, Krüger M, Wang ZQ, Platzer M, Englert C
Hum Mol Genet 2017, 26(9), 1694-705 - Die Bibliothek im Körper
Neri F
GIT Labor-Fachzeitschrift 2017, 8, 14-7 - Intragenic DNA methylation prevents spurious transcription initiation.
Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S
Nature 2017, 543(7643), 72-7 - Alternative splicing of SMPD1 coding for acid sphingomyelinase in major depression.
Rhein C, Reichel M, Kramer M, Rotter A, Lenz B, Mühle C, Gulbins E, Kornhuber J
J Affect Disord 2017, 209, 10-5 - Massive Effect on LncRNAs in Human Monocytes During Fungal and Bacterial Infections and in Response to Vitamins A and D.
Riege K, Hölzer M, Klassert TE, Barth E, Bräuer J, Collatz M, Hufsky F, Mostajo N, Stock M, Vogel B, Slevogt H, Marz M
Sci Rep 2017, 7, 40598 - MicroRNA miR-29 controls a compensatory response to limit neuronal iron accumulation during adult life and aging.
Ripa R, Dolfi L, Terrigno M, Pandolfini L, Savino A, Arcucci V, Groth M, Terzibasi Tozzini E, Baumgart M, Cellerino A
BMC Biol 2017, 15(1), 9 - Positive Selektion in langlebigen Nagern und kurzlebigen Fischen : eine bioinformatische Suche nach der genetischen Basis des Alterns
Sahm A
Dissertation 2017, Jena, Germany - Parallel evolution of genes controlling mitonuclear balance in short-lived annual fishes.
Sahm A, Bens M, Platzer M, Cellerino A
Aging Cell 2017, 16(3), 488-96 - PosiGene: automated and easy-to-use pipeline for genome-wide detection of positively selected genes.
Sahm A, Bens M, Platzer M, Szafranski K
Nucleic Acids Res 2017, 45(11), e100