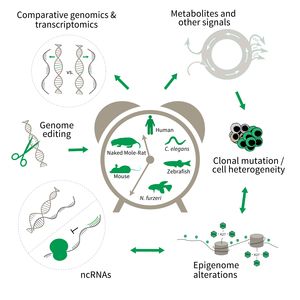
Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influ ence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2018
- Googles DeepVariant: eine Methode für die Medizin- und Bioinformatik?
Fürstberger** A, Platzer** M, Kestler** HA
BIOspektrum 2018, 24(3), 235–235 ** co-corresponding authors - Effects of allelic variations in the human Myxovirus resistance protein A on its antiviral activity.
Graf L, Dick A, Sendker F, Barth E, Marz M, Daumke O, Kochs G
J Biol Chem 2018, 293(9), 3056-72 - Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging.
Heinze* I, Bens* M, Calzia* E, Holtze S, Dakhovnik O, Sahm A, Kirkpatrick JM, Szafranski K, Romanov N, Sama SN, Holzer K, Singer S, Ermolaeva M, Platzer** M, Hildebrandt** T, Ori** A
BMC Biol 2018, 16(1), 82 * equal contribution, ** co-senior authors - Multiple roots of fruiting body formation in Amoebozoa.
Hillmann F, Forbes G, Novohradská S, Ferling I, Riege K, Groth M, Westermann M, Marz M, Spaller T, Winckler T, Schaap P, Glöckner G
Genome Biol Evol 2018, 10(2), 591-606 - The microenvironment of naked mole-rat burrows in East Africa
Holtze S, Braude S, Lemma A, Koch R, Morhart M, Szafranski K, Platzer M, Alemayehu F, Goeritz F, Hildebrandt TB
Afr J Ecol 2018, 56(2), 279-89 - 3D Network exploration and visualisation for lifespan data.
Hühne* R, Kessler* V, Fürstberger* A, Kühlwein S, Platzer M, Sühnel J, Lausser L, Kestler HA
BMC Bioinformatics 2018, 19(1), 390 * equal contribution - Bioinformatics Meets Virology: The European Virus Bioinformatics Center's Second Annual Meeting.
Ibrahim B, Arkhipova K, Andeweg AC, Posada-Céspedes S, Enault F, Gruber A, Koonin EV, Kupczok A, Lemey P, McHardy AC, McMahon DP, Pickett BE, Robertson DL, Scheuermann RH, Zhernakova A, Zwart MP, Schönhuth A, Dutilh BE, Marz M
Viruses 2018, 10(5, 256; https://doi.org/10.33 - Epigenetic Erosion in Adult Stem Cells: Drivers and Passengers of Aging.
Kosan C, Heidel FH, Godmann M, Bierhoff H
Cells 2018, 7(12) - Identification of potential microRNAs associated with Herpesvirus family based on bioinformatic analysis.
Lamkiewicz K, Barth E, Marz** M, Ibrahim** B
bioRxiv 2018, https://doi.org/10.1101/417782 ** co-corresponding authors - Wilms Tumor 1b defines a wound-specific sheath cell subpopulation associated with notochord repair.
Lopez-Baez JC, Simpson DJ, LLeras Forero L, Zeng Z, Brunsdon H, Salzano A, Brombin A, Wyatt C, Rybski W, Huitema LFA, Dale RM, Kawakami K, Englert C, Chandra T, Schulte-Merker S, Hastie ND, Patton EE
Elife 2018, 7, e30657