Subarea 3: Genetics and Epigenetics of Aging
The focus of Subarea 3 is on genetic and epigenetic determinants of life- and health span as well as aging in fish, rodents and humans. This line of research builds on the expertise of the institute in comparative and functional genomics.
The research is defined by five focus areas:
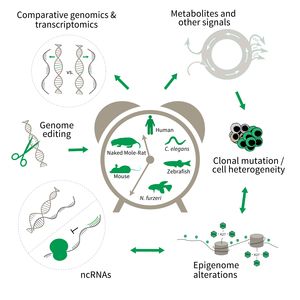
- Comparative genomics in short- and long-lived models of aging,
- Genomic engineering in N. furzeri,
- Epigenetics of aging,
- Non-coding RNAs in aging, and
- Comparative transcriptomics of aging.
Research focus of Subarea 3.
To uncover causative factors for aging, comparative genomics in short- and long-lived model systems are applied. Functional genomics is used to identify novel pathways contribute to aging of an organism and to validate the functional relevance of genetic and epigenetic changes that occur during aging. Furthermore, genetic risk factors for aging-related diseases are identified and functionally tested. The future development of the Subarea aims to integrate changes in host-microbiota interactions during aging, and how these influence clonal mutation and epigenetic alterations through metabolites and other signals.
Publications
(since 2016)
2019
- Wilms Tumor 1b Expression Defines a Pro-regenerative Macrophage Subtype and Is Required for Organ Regeneration in the Zebrafish.
Sanz-Morejón A, García-Redondo AB, Reuter H, Marques IJ, Bates T, Galardi-Castilla M, Große A, Manig S, Langa X, Ernst A, Piragyte I, Botos MA, González-Rosa JM, Ruiz-Ortega M, Briones AM, Salaices M, Englert C, Mercader N
Cell Rep 2019, 28(5), 1296-1306.e6 - Author Correction: Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals.
Schwörer S, Becker F, Feller C, Baig AH, Köber U, Henze H, Kraus JM, Xin B, Lechel A, Lipka DB, Varghese CS, Schmidt M, Rohs R, Aebersold R, Medina KL, Kestler HA, Neri F, von Maltzahn** J, Tümpel** S, Rudolph** KL
Nature 2019, 572(7769), E11-5 ** co-corresponding authors - The landscape of the alternatively spliced transcriptome remains stable during aging across different species and tissues.
Sieber P, Barth* E, Marz* M
bioRxiv 2019, https://doi.org/10.1101/541417 * equal contribution - Tissue-specific Gene Expression Changes Are Associated With Aging In Mice
Srivastava A, Barth E, Ermolaeva M, Guenther M, Frahm C, Marz M, Witte OW
Genomics Proteomics Bioinformatics 2019 - Direct RNA nanopore sequencing of full-length coronavirus genomes provides novel insights into structural variants and enables modification analysis.
Viehweger* A, Krautwurst* S, Lamkiewicz K, Madhugiri R, Ziebuhr J, Hölzer M, Marz M
Genome Res 2019, 29(9), 1545-54 * equal contribution - Erratum for Wegner et al., "Biogeochemical Regimes in Shallow Aquifers Reflect the Metabolic Coupling of the Elements Nitrogen, Sulfur, and Carbon".
Wegner CE, Gaspar M, Geesink P, Herrmann M, Marz M, Küsel K
Appl Environ Microbiol 2019, 85(9) - Biogeochemical regimes in shallow aquifers reflect the metabolic coupling of elements of nitrogen, sulfur and carbon.
Wegner CE, Gaspar M, Geesink P, Herrmann M, Marz M, Küsel K
Appl Environ Microbiol 2019, 85(5), pii: e02346-18
2018
- Cell-based RNAi screening and high-content analysis in primary calvarian osteoblasts applied to identification of osteoblast differentiation regulators.
Ahmad M, Kroll T, Jakob J, Rauch A, Ploubidou A, Tuckermann J
Sci Rep 2018, 8(1), 14045 - Cell cycle-dependent and -independent telomere shortening accompanies murine brain aging.
Ain Q, Schmeer C, Penndorf D, Fischer M, Bondeva T, Förster M, Haenold R, Witte OW, Kretz A
Aging (Albany NY) 2018, 10(11), 3397-420 - Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly.
Aramillo Irizar P, Schäuble S, Esser D, Groth M, Frahm C, Priebe S, Baumgart M, Hartmann N, Marthandan S, Menzel U, Müller J, Schmidt S, Ast V, Caliebe A, König R, Krawczak M, Ristow M, Schuster S, Cellerino A, Diekmann S, Englert C, Hemmerich P, Sühnel J, Guthke R, Witte OW, Platzer M, Ruppin E, Kaleta C
Nat Commun 2018, 9(1), 327