Subarea 4: Cell Dynamics and Molecular Damages in Aging
The research focus of Subarea 4 is on studying damages of macromolecules (proteins, nucleic acids) and determining the structure-function relationship of biomolecules relevant to damage and damage repair processes and responses to molecular damage that might lead to aging and aging-associated pathologies.
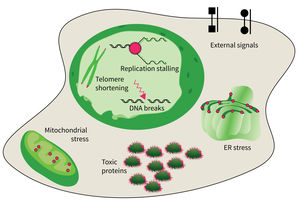
The studies are focused on the following research areas: DNA replication, DNA damage responses (DDR), stress responses, metabolic stresses, protein trafficking and protein damages.
The research is defined by four focus areas:
- DNA damage response in tissue homeostasis and neuropathies,
- Quality control in the endoplasmic reticulum for secretory pathway in aging processes,
- Intrinsic and extrinsic factors implicated in cellular decline during aging, and
- DNA replication and genomic integrity preventing premature aging and diseases.
Research focus of Subarea 4.
The accumulation of damaged macromolecules or subcellular organelles is associated with dysfunction of a cell, which contributes to tissue & organ failure. DNA damage, genomic instability, protein misfolding or defects in toxic protein degradation can compromise cell functionality. Alterations of mitochondrial DNA and protein complexes affect cellular metabolism, which will have a general impact on cell integrity.
Publications
(since 2016)
2019
- Design, synthesis and characterization of new macrocyclic inhibitors of the proprotein convertase furin.
Lam van TV, Ivanova T, Hardes K, Heindl MR, Morty RE, Böttcher-Friebertshäuser E, Lindberg I, Than ME, Dahms SO, Steinmetzer T
ChemMedChem 2019, 14(6), 673-85 - The role of the anti-aging protein klotho in the brain
Rothe M
Dissertation 2019, Jena, Germany - Local inhibition of rRNA transcription without nucleolar segregation after targeted ion irradiation of the nucleolus.
Siebenwirth C, Greubel C, Drexler GA, Reindl J, Walsh DWM, Schwarz B, Sammer M, Baur I, Pospiech H, Schmid TE, Dollinger G, Friedl AA
J Cell Sci 2019, 132(19), jcs232181 - Tissue-specific Gene Expression Changes Are Associated With Aging In Mice
Srivastava A, Barth E, Ermolaeva M, Guenther M, Frahm C, Marz M, Witte OW
Genomics Proteomics Bioinformatics 2019 - Tmem30a is essential for the membrane raft assembly and EpoR signal transduction in erythropoiesis
Yang F
Dissertation 2019, Jena, Germany
2018
- Klotho expression is a prerequisite for proper muscle stem cell function and regeneration of skeletal muscle.
Ahrens* HE, Huettemeister* J, Schmidt M, Kaether C, von Maltzahn J
Skelet Muscle 2018, 8(1), 20 * equal contribution - Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly.
Aramillo Irizar P, Schäuble S, Esser D, Groth M, Frahm C, Priebe S, Baumgart M, Hartmann N, Marthandan S, Menzel U, Müller J, Schmidt S, Ast V, Caliebe A, König R, Krawczak M, Ristow M, Schuster S, Cellerino A, Diekmann S, Englert C, Hemmerich P, Sühnel J, Guthke R, Witte OW, Platzer M, Ruppin E, Kaleta C
Nat Commun 2018, 9(1), 327 - X-ray Structures of the Proprotein Convertase Furin Bound with Substrate Analogue Inhibitors Reveal Substrate Specificity Determinants beyond the S4 Pocket.
Dahms SO, Hardes K, Steinmetzer T, Than ME
Biochemistry 2018, 57(6), 925-34 - Raman and infrared spectroscopy reveal that proliferating and quiescent human fibroblast cells age by biochemically similar but not identical processes.
Eberhardt K, Matthäus C, Marthandan S, Diekmann* S, Popp* J
PLoS One 2018, 13(12), e0207380 * equal contribution - Cellular and epigenetic drivers of stem cell ageing.
Ermolaeva** M, Neri** F, Ori** A, Rudolph** KL
Nat Rev Mol Cell Biol 2018, 19(9), 594-610 ** co-corresponding authors