Subarea 4: Cell Dynamics and Molecular Damages in Aging
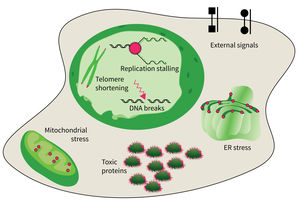
The research focus of Subarea 4 is on studying damages of macromolecules (proteins, nucleic acids) and determining the structure-function relationship of biomolecules relevant to damage and damage repair processes and responses to molecular damage that might lead to aging and aging-associated pathologies.
The studies are focused on the following research areas: DNA replication, DNA damage responses (DDR), stress responses, metabolic stresses, protein trafficking and protein damages.
The research is defined by four focus areas:
- DNA damage response in tissue homeostasis and neuropathies,
- Quality control in the endoplasmic reticulum for secretory pathway in aging processes,
- Intrinsic and extrinsic factors implicated in cellular decline during aging, and
- DNA replication and genomic integrity preventing premature aging and diseases.
Research focus of Subarea 4.
The accumulation of damaged macromolecules or subcellular organelles is associated with dysfunction of a cell, which contributes to tissue & organ failure. DNA damage, genomic instability, protein misfolding or defects in toxic protein degradation can compromise cell functionality. Alterations of mitochondrial DNA and protein complexes affect cellular metabolism, which will have a general impact on cell integrity.
Publications
(since 2016)
2023
- Impact of Hypermannosylation on the Structure and Functionality of the ER and the Golgi Complex.
Franzka P, Schüler SC, Kentache T, Storm R, Bock A, Katona I, Weis J, Buder K, Kaether C, Hübner CA
Biomedicines 2023, 11(1), 146.doi: 10.3390/biomedicines110 - The biological functions of PARP1
Gong Y
Dissertation 2023, Jena, Germany - Separation-of-function study of PARP1
Kamaletdinova T
Dissertation 2023, Jena, Germany - Poly(ADP-Ribose) Polymerase-1 Lacking Enzymatic Activity Is Not Compatible with Mouse Development.
Kamaletdinova T, Zong W, Urbánek P, Wang S, Sannai M, Grigaravičius P, Sun W, Fanaei-Kahrani Z, Mangerich A, Hottiger MO, Li T, Wang ZQ
Cells 2023, 12(16), 2078 - Dissecting the UV-B effect on adaptive responses through changes in mitochondrial homeostasis
Martirosyan A
Dissertation 2023, Jena, Germany - Protein quality control: from molecular mechanisms to therapeutic intervention-EMBO workshop, May 21-26 2023, Srebreno, Croatia.
Münch C, Kirstein J
Cell Stress Chaperones 2023, 28(6), 631-40 - DNA damage response in neurodevelopment and neuromaintenance.
Qing* X, Zhang* G, Wang ZQ
FEBS J 2023, 290(13), 3300-10 * equal contribution - Simultaneous Nbs1 and p53 inactivation in neural progenitors triggers High-Grade Gliomas (HGG).
Reuss DE, Downing SM, Camacho CV, Wang YD, Piro RM, Herold-Mende C, Wang ZQ, Hofmann TG, Sahm F, von Deimling A, McKinnon PJ, Frappart PO
Neuropathol Appl Neurobiol 2023, 49(4), e12915 - ABRAXAS1 orchestrates BRCA1 activities to counter genome destabilizing repair pathways-lessons from breast cancer patients.
Sachsenweger J, Jansche R, Merk T, Heitmeir B, Deniz M, Faust U, Roggia C, Tzschach A, Schroeder C, Riess A, Pospiech H, Peltoketo H, Pylkäs K, Winqvist R, Wiesmüller L
Cell Death Dis 2023, 14(5), 328 - Abl depletion via autophagy mediates the beneficial effects of quercetin against Alzheimer pathology across species.
Schiavi A, Cirotti C, Gerber LS, Di Lauro G, Maglioni S, Shibao PYT, Montresor S, Kirstein J, Petzsch P, Köhrer K, Schins RPF, Wahle T, Barilà D, Ventura N
Cell Death Discov 2023, 9(1), 376