Subarea 4: Cell Dynamics and Molecular Damages in Aging
The research focus of Subarea 4 is on studying damages of macromolecules (proteins, nucleic acids) and determining the structure-function relationship of biomolecules relevant to damage and damage repair processes and responses to molecular damage that might lead to aging and aging-associated pathologies.
The studies are focused on the following research areas: DNA replication, DNA damage responses (DDR), stress responses, metabolic stresses, protein trafficking and protein damages.
The research is defined by four focus areas:
- DNA damage response in tissue homeostasis and neuropathies,
- Quality control in the endoplasmic reticulum for secretory pathway in aging processes,
- Intrinsic and extrinsic factors implicated in cellular decline during aging, and
- DNA replication and genomic integrity preventing premature aging and diseases.
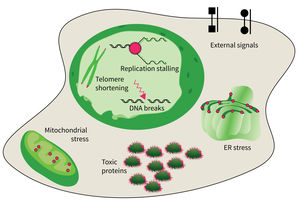
Research focus of Subarea 4.
The accumulation of damaged macromolecules or subcellular organelles is associated with dysfunction of a cell, which contributes to tissue & organ failure. DNA damage, genomic instability, protein misfolding or defects in toxic protein degradation can compromise cell functionality. Alterations of mitochondrial DNA and protein complexes affect cellular metabolism, which will have a general impact on cell integrity.
Publications
(since 2016)
2017
- The protein-tyrosine phosphatase DEP-1 promotes migration and phagocytic activity of microglial cells in part through negative regulation of fyn tyrosine kinase.
Schneble N, Müller J, Kliche S, Bauer R, Wetzker R, Böhmer FD, Wang ZQ, Müller JP
Glia 2017, 65(2), 416-28 - PARPing for Balance in the Homeostasis of Poly(ADP-ribosyl)ation.
Schuhwerk H, Atteya R, Siniuk K, Wang ZQ
Semin Cell Dev Biol 2017, 63, 81-91 - Kinetics of poly(ADP-ribosyl)ation, but not PARP1 itself, determines the cell fate in response to DNA damage in vitro and in vivo.
Schuhwerk H, Bruhn C, Siniuk K, Min W, Erener S, Grigaravicius P, Krüger A, Ferrari E, Zubel T, Lazaro D, Monajembashi S, Kiesow K, Kroll T, Bürkle A, Mangerich A, Hottiger M, Wang ZQ
Nucleic Acids Res 2017, 45(19), 11174-92 - Local responses of S phase cells post irradiation damage
Smith SJ
Dissertation 2017, Jena, Germany - Cdc45-induced loading of human RPA onto single-stranded DNA.
Szambowska A, Tessmer I, Prus P, Schlott B, Pospiech H, Grosse F
Nucleic Acids Res 2017, 45(6), 3217-30 - Lysine Acetylation and Deacetylation in Brain Development and Neuropathies.
Tapias A, Wang ZQ
Genomics Proteomics Bioinformatics 2017, 15(1), 19-36 - TRH action is impaired in pituitaries of male IGSF1-deficient mice.
Turgeon MO, Silander TL, Doycheva D, Liao XH, Rigden M, Ongaro L, Zhou X, Joustra SD, Wit JM, Wade MG, Heuer H, Refetoff S, Bernard DJ
Endocrinology 2017, 158(4), 815-30 - The sorting receptor Rer1 controls Purkinje cell function via voltage gated sodium channels.
Valkova C, Liebmann L, Krämer A, Hübner CA, Kaether C
Sci Rep 2017, 7, 41248
2016
- Differential sensitivity of lipegfilgrastim and pegfilgrastim to neutrophil elastase correlates with differences in clinical pharmacokinetic profile.
Abdolzade-Bavil A, von Kerczek A, Cooksey BA, Kaufman T, Krasney PA, Pukac L, Görlach M, Lammerich A, Scheckermann C, Allgaier H, Shen WD, Liu PM
J Clin Pharmacol 2016, 56(2), 186-94 - Heme interacts with histidine- and tyrosine-based protein motifs and inhibits enzymatic activity of chloramphenicol acetyltransferase from Escherichiacoli.
Brewitz HH, Goradia N, Schubert E, Galler K, Kühl T, Syllwasschy B, Popp J, Neugebauer U, Hagelueken G, Schiemann O, Ohlenschläger** O, Imhof** D
Bba-Gen Subjects 2016, 1860(6), 1343-53 ** co-corresponding authors