Subarea 4: Cell Dynamics and Molecular Damages in Aging
The research focus of Subarea 4 is on studying damages of macromolecules (proteins, nucleic acids) and determining the structure-function relationship of biomolecules relevant to damage and damage repair processes and responses to molecular damage that might lead to aging and aging-associated pathologies.
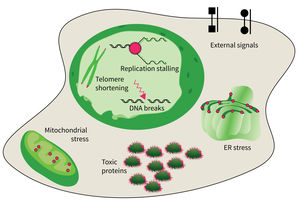
The studies are focused on the following research areas: DNA replication, DNA damage responses (DDR), stress responses, metabolic stresses, protein trafficking and protein damages.
The research is defined by four focus areas:
- DNA damage response in tissue homeostasis and neuropathies,
- Quality control in the endoplasmic reticulum for secretory pathway in aging processes,
- Intrinsic and extrinsic factors implicated in cellular decline during aging, and
- DNA replication and genomic integrity preventing premature aging and diseases.
Research focus of Subarea 4.
The accumulation of damaged macromolecules or subcellular organelles is associated with dysfunction of a cell, which contributes to tissue & organ failure. DNA damage, genomic instability, protein misfolding or defects in toxic protein degradation can compromise cell functionality. Alterations of mitochondrial DNA and protein complexes affect cellular metabolism, which will have a general impact on cell integrity.
Publications
(since 2016)
2017
- Optimization of Substrate-Analogue Furin Inhibitors.
Ivanova T, Hardes K, Kallis S, Dahms SO, Than ME, Künzel S, Böttcher-Friebertshäuser E, Lindberg I, Jiao GS, Bartenschlager R, Steinmetzer T
ChemMedChem 2017, 12(23), 1953-68 - Regulation und Deregulation eukaryotischer DNA-Replikation
Keller-Koalick D
Dissertation 2017, Jena, Germany - The E3 ubiquitin ligase APC/C(C)(dh1) degrades MCPH1 after MCPH1-βTrCP2-Cdc25A-mediated mitotic entry to ensure neurogenesis.
Liu X, Zong W, Li T, Wang Y, Xu X, Zhou** ZW, Wang** ZQ
EMBO J 2017, 36(24), 3666-81 ** co-corresponding authors - Assessment of HDACi-Induced Cytotoxicity.
Marx-Blümel L, Marx C, Kühne M, Sonnemann J
Methods Mol Biol 2017, 1510, 23-45 - Telomerase abrogates aneuploidy-induced telomere replication stress, senescence and cell depletion.
Meena JK, Cerutti A, Beichler C, Morita Y, Bruhn C, Kumar M, Kraus JM, Speicher MR, Wang ZQ, Kestler HA, Fagagna Fdd, Günes C, Rudolph KL
EMBO J 2017, 36(19), 2922-4 Erratum for EMBO J 2015 volume 34 page 1371 - The Wilms tumor protein Wt1 contributes to female fertility by regulating oviductal proteostasis.
Nathan A, Reinhardt P, Kruspe D, Jörß T, Groth M, Nolte H, Habenicht A, Herrmann J, Holschbach V, Toth B, Krüger M, Wang ZQ, Platzer M, Englert C
Hum Mol Genet 2017, 26(9), 1694-705 - DNA damage in protective and adverse inflammatory responses: Friend of foe?
Poliezhaieva T, Ermolaeva M
Mech Ageing Dev 2017, 165(Pt A), 47-53 - N-Heterocyclic (4-Phenylpiperazin-1-yl)methanones Derived from Phenoxazine and Phenothiazine as Highly Potent Inhibitors of Tubulin Polymerization.
Prinz H, Ridder AK, Vogel K, Böhm KJ, Ivanov I, Ghasemi J, Aghaee E, Müller K:
J Med Chem 2017, 60(2), 749-66 - Central immune tolerance depends on crosstalk between the classical and alternative NF-κB pathways in medullary thymic epithelial cells.
Riemann M, Andreas N, Fedoseeva M, Meier E, Weih D, Freytag H, Schmidt-Ullrich R, Klein U, Wang ZQ, Weih F
J Autoimmun 2017, 81, 56-67 - The protein-tyrosine phosphatase DEP-1 promotes migration and phagocytic activity of microglial cells in part through negative regulation of fyn tyrosine kinase.
Schneble N, Müller J, Kliche S, Bauer R, Wetzker R, Böhmer FD, Wang ZQ, Müller JP
Glia 2017, 65(2), 416-28