Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
Research focus of Subarea 5.
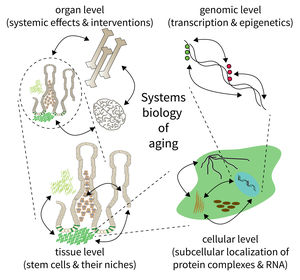
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2023
- Impact of Hypermannosylation on the Structure and Functionality of the ER and the Golgi Complex.
Franzka P, Schüler SC, Kentache T, Storm R, Bock A, Katona I, Weis J, Buder K, Kaether C, Hübner CA
Biomedicines 2023, 11(1), 146.doi: 10.3390/biomedicines110 - Comparative analysis of thyroid hormone systems in rodents with subterranean lifestyle.
Gerhardt P, Begall S, Frädrich C, Renko K, Hildebrandt TB, Holtze S, Heinrich A, Sahm A, Meci X, Köhrle J, Rijntjes E, Henning Y
Sci Rep 2023, 13(1), 3122 - Author Correction: The evolutionary history of 2,658 cancers.
Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, Mitchell TJ, Rubanova Y, Anur P, Yu K, Tarabichi M, Deshwar A, Wintersinger J, Kleinheinz K, Vázquez-García I, Haase K, Jerman L, Sengupta S, Macintyre G, Malikic S, Donmez N, Livitz DG, Cmero M, Demeulemeester J, Schumacher S, Fan Y, Yao X, Lee J, Schlesner M, Boutros PC, Bowtell DD, Zhu H, Getz G, Imielinski M, Beroukhim R, Sahinalp SC, Ji Y, Peifer M, Markowetz F, Mustonen V, Yuan K, Wang W, Morris QD, PCAWG Evolution and Heterogeneity Working Group, Spellman PT, Wedge DC, Van Loo P, PCAWG Consortium
Nature 2023, 614(7948), E42 - Liquid biopsy: an examination of platelet RNA obtained from head and neck squamous cell carcinoma patients for predictive molecular tumor markers.
Huber* LT, Kraus* JM, Ezić* J, Wanli A, Groth M, Laban S, Hoffmann TK, Wollenberg B, Kestler** HA, Brunner** C
Explor Target Antitumor Ther 2023, 4(3), 422–446 * equal contribution, ** co-corresponding authors - Author Correction: Pan-cancer analysis of whole genomes.
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium
Nature 2023, 614(7948), E39 - Evolutionary paths to mammalian longevity through the lens of gene expression.
Işıldak U, Dönertaş HM
EMBO J 2023, 42(17), e114879 - Membrane shapers from two distinct superfamilies cooperate in the development of neuronal morphology.
Izadi M, Wolf D, Seemann E, Ori A, Schwintzer L, Steiniger F, Kessels MM, Qualmann B
J Cell Biol 2023, 222(8), e202211032 - Crafting figures to describe cellular processes.
Kmiecik** SW, Shi** Q, Chen** YG, Moormann** J, Hildebrandt** TM, Schade** A, Fischer** M, Kim** KQ
Trends Biochem Sci 2023, 48(6), 491-4 ** co-corresponding authors - Author Correction: Patterns of somatic structural variation in human cancer genomes.
Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE, Imielinski M, PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Campbell PJ, PCAWG Consortium
Nature 2023, 614(7948), E38 - Focal structural variants revealed by whole genome sequencing disrupt the histone demethylase KDM4C in B cell lymphomas.
Lopez C, Schleussner N, Bernhart SH, Kleinheinz K, Sungalee S, Sczakiel HL, Kretzmer H, Toprak UH, Glaser S, Wagener R, Ammerpohl O, Bens S, Giefing M, Sanchez JCG, Apic G, Hubschmann D, Janz M, Kreuz M, Mottok A, Muller JM, Seufert J, Hoffmann S, Korbel JO, Russell RB, Schule R, Trumper L, Klapper W, Radlwimmer B, Lichter P, Kuppers R, Schlesner M, Mathas S, Siebert R
Haematologica 2023, 108(2), 543-54