Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
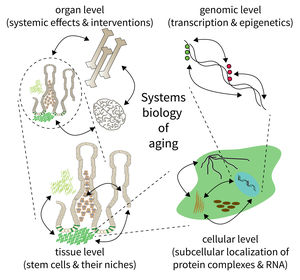
Research focus of Subarea 5.
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2018
- DIEGO: Detection of Differential Alternative Splicing using Aitchison's Geometry.
Doose G, Bernhart SH, Wagener R, Hoffmann S
Bioinformatics 2018, 34(6), 1066-8 - Author Correction: TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells.
Elster D, Tollot M, Schlegelmilch K, Ori A, Rosenwald A, Sahai E, von Eyss B
Nat Commun 2018, 9(1), 3781 Erratum for Nat Commun. 2018 volume 9 page 3115 - TRPS1 shapes YAP/TEAD-dependent transcription in breast cancer cells.
Elster* D, Tollot* M, Schlegelmilch K, Ori A, Rosenwald A, Sahai E, von Eyss B
Nat Commun 2018, 9(1), 3115 * equal contribution - Cellular and epigenetic drivers of stem cell ageing.
Ermolaeva** M, Neri** F, Ori** A, Rudolph** KL
Nat Rev Mol Cell Biol 2018, 19(9), 594-610 ** co-corresponding authors - Googles DeepVariant: eine Methode für die Medizin- und Bioinformatik?
Fürstberger** A, Platzer** M, Kestler** HA
BIOspektrum 2018, 24(3), 235–235 ** co-corresponding authors - Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging.
Heinze* I, Bens* M, Calzia* E, Holtze S, Dakhovnik O, Sahm A, Kirkpatrick JM, Szafranski K, Romanov N, Sama SN, Holzer K, Singer S, Ermolaeva M, Platzer** M, Hildebrandt** T, Ori** A
BMC Biol 2018, 16(1), 82 * equal contribution, ** co-senior authors - 3D Network exploration and visualisation for lifespan data.
Hühne* R, Kessler* V, Fürstberger* A, Kühlwein S, Platzer M, Sühnel J, Lausser L, Kestler HA
BMC Bioinformatics 2018, 19(1), 390 * equal contribution - The Influence of Multi-class Feature Selection on the Prediction of Diagnostic Phenotypes
Lausser L, Szekely R, Schirra LR, Kestler HA
Neural Process Lett 2018, 48(2), 863–880 - Rank-based classifiers for extremely high-dimensional gene expression data
Lausser* L, Schmid* F, Schirra* LR, Wilhelm AFX, Kestler HA
ADV DATA ANAL CLASSI 2018, 12(4), 917–936 * equal contribution - Quantifying compartment-associated variations of protein abundance in proteomics data.
Parca L, Beck M, Bork P, Ori A
Mol Syst Biol 2018, 14(7), e8131