Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
Research focus of Subarea 5.
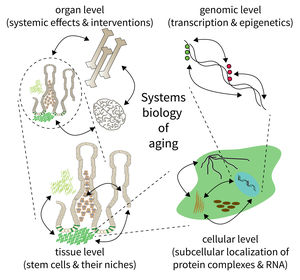
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2019
- Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma.
López C, Kleinheinz K, Aukema SM, Rohde M, Bernhart SH, Hübschmann D, Wagener R, Toprak UH, Raimondi F, Kreuz M, Waszak SM, Huang Z, Sieverling L, Paramasivam N, Seufert J, Sungalee S, Russell RB, Bausinger J, Kretzmer H, Ammerpohl O, Bergmann AK, Binder H, Borkhardt A, Brors B, Claviez A, Doose G, Feuerbach L, Haake A, Hansmann ML, Hoell J, Hummel M, Korbel JO, Lawerenz C, Lenze D, Radlwimmer B, Richter J, Rosenstiel P, Rosenwald A, Schilhabel MB, Stein H, Stilgenbauer S, Stadler PF, Szczepanowski M, Weniger MA, Zapatka M, Eils R, Lichter P, Loeffler M, Möller P, Trümper L, Klapper W, ICGC MMML-Seq Consortium, Hoffmann S, Küppers R, Burkhardt B, Schlesner M, Siebert R
Nat Commun 2019, 10(1), 1459 - Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time.
Muntel* J, Kirkpatrick* J, Bruderer R, Huang T, Vitek O, Ori** A, Reiter** L
J Proteome Res 2019, 18(3), 1340-51 * equal contribution, ** co-senior authors - Author Correction: Metastatic-niche labelling reveals parenchymal cells with stem features.
Ombrato L, Nolan E, Kurelac I, Mavousian A, Bridgeman VL, Heinze I, Chakravarty P, Horswell S, Gonzalez-Gualda E, Matacchione G, Weston A, Kirkpatrick J, Husain E, Speirs V, Collinson L, Ori A, Lee JH, Malanchi I
Nature 2019, 575(7784), E8 - Metastatic-niche labelling reveals parenchymal cells with stem features.
Ombrato L, Nolan E, Kurelac I, Mavousian A, Bridgeman VL, Heinze I, Chakravarty P, Horswell S, Gonzalez-Gualda E, Matacchione G, Weston A, Kirkpatrick J, Husain E, Speirs V, Collinson L, Ori A, Lee JH, Malanchi I
Nature 2019, 572(7771), 603-8 - Disentangling Genetic and Environmental Effects on the Proteotypes of Individuals.
Romanov N, Kuhn M, Aebersold R, Ori A, Beck M, Bork P
Cell 2019, 177(5), 1308-18 - RB, p130 and p107 differentially repress G1/S and G2/M genes after p53 activation.
Schade AE, Fischer M, DeCaprio JA
Nucleic Acids Res 2019, 47(21), 11197-208 - Author Correction: Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals.
Schwörer S, Becker F, Feller C, Baig AH, Köber U, Henze H, Kraus JM, Xin B, Lechel A, Lipka DB, Varghese CS, Schmidt M, Rohs R, Aebersold R, Medina KL, Kestler HA, Neri F, von Maltzahn** J, Tümpel** S, Rudolph** KL
Nature 2019, 572(7769), E11-5 ** co-corresponding authors - Increased Expression of Immature Mannose-Containing Glycoproteins and Sialic Acid in Aged Mouse Brains.
Simon F, Bork K, Gnanapragassam VS, Baldensperger T, Glomb MA, Di Sanzo S, Ori A, Horstkorte R
Int J Mol Sci 2019, 20(24), 6118 - Gene expression profile of human T cells following a single stimulation of peripheral blood mononuclear cells with anti-CD3 antibodies.
Sousa IG, Simi KCR, do Almo MM, Bezerra MAG, Doose G, Raiol T, Stadler PF, Hoffmann S, Maranhão AQ, Brigido MM
BMC Genomics 2019, 20(1), 593 - DREAM and RB cooperate to induce gene repression and cell-cycle arrest in response to p53 activation.
Uxa S, Bernhart SH, Mages CFS, Fischer M, Kohler R, Hoffmann S, Stadler PF, Engeland K, Müller GA
Nucleic Acids Res 2019, 47(17), 9087-9103.