Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
Research focus of Subarea 5.
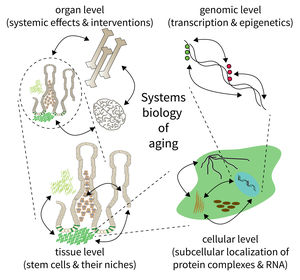
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2021
- Alternative Animal Models of Aging Research
Holtze S, Gorshkova E, Braude S, Cellerino A, Dammann P, Hildebrandt TB, Hoeflich A, Hoffmann S, Koch P, Terzibasi Tozzini E, Skulachev M, Skulachev VP, Sahm A
Front Mol Biosci 2021, 8, doi: 10.3389/fmolb.2021.660959. - Mutational mechanisms shaping the coding and noncoding genome of germinal center derived B-cell lymphomas.
Hübschmann D, Kleinheinz K, Wagener R, Bernhart SH, López C, Toprak UH, Sungalee S, Ishaque N, Kretzmer H, Kreuz M, Waszak SM, Paramasivam N, Ammerpohl O, Aukema SM, Beekman R, Bergmann AK, Bieg M, Binder H, Borkhardt A, Borst C, Brors B, Bruns P, Carrillo de Santa Pau E, Claviez A, Doose G, Haake A, Karsch D, Haas S, Hansmann ML, Hoell JI, Hovestadt V, Huang B, Hummel M, Jäger-Schmidt C, Kerssemakers JNA, Korbel JO, Kube D, Lawerenz C, Lenze D, Martens JHA, Ott G, Radlwimmer B, Reisinger E, Richter J, Rico D, Rosenstiel P, Rosenwald A, Schillhabel M, Stilgenbauer S, Stadler PF, Martín-Subero JI, Szczepanowski M, Warsow G, Weniger MA, Zapatka M, Valencia A, Stunnenberg HG, Lichter P, Möller P, Loeffler M, Eils R, Klapper W, Hoffmann S, Trümper L, ICGC MMML-Seq consortium, ICGC DE-Mining consortium, BLUEPRINT consortium, Küppers R, Schlesner M, Siebert R
Leukemia 2021, 35(7), 2002-16 published during change of institution - Fetal-like reversion in the regenerating intestine is regulated by mesenchymal Asporin
Iqbal S, Andersson S, Nestaite E, Pentinmikko N, Kumar A, Borshagovski D, Webb A, Saarinen T, Juuti A, Ori A, Varjosalo M, Pietiläinen KH, Jensen KB, Oudhoff M, Katajisto P
bioRxiv 2021, https://doi.org/10.1101/2021.06. - ATR regulates neuronal activity by modulating presynaptic firing.
Kirtay M, Sell J, Marx C, Haselmann H, Ceanga M, Zhou ZW, Rahmati V, Kirkpatrick J, Buder K, Grigaravicius P, Ori A, Geis** C, Wang** ZQ
Nat Commun 2021, 12(1), 4067 ** co-corresponding authors - Simultaneous expression of MMB-FOXM1 complex components enables efficient bypass of senescence.
Kumari R, Hummerich* H, Shen* X, Fischer* M, Litovchick L, Mittnacht S, DeCaprio JA, Jat PS
Sci Rep 2021, 11(1), 21506 * equal contribution - The N-terminal BRCT domain determines MCPH1 function in brain development and fertility.
Liu* X, Schneble-Löhnert* N, Kristofova M, Qing X, Labisch J, Hofmann S, Ehrenberg S, Sannai M, Jörß T, Ori A, Godmann M, Wang ZQ
Cell Death Dis 2021, 12(2), 143 * equal contribution - Implementing FAIR data management within the German Network for Bioinformatics Infrastructure (de.NBI) exemplified by selected use cases.
Mayer* G, Müller* W, Schork K, Uszkoreit J, Weidemann A, Wittig U, Rey M, Quast C, Felden J, Glöckner FO, Lange M, Arend D, Beier S, Junker A, Scholz U, Schüler D, Kestler HA, Wibberg D, Pühler A, Twardziok S, Eils J, Eils R, Hoffmann S, Eisenacher M, Turewicz M
Brief Bioinform 2021, 22(5), bbab010 * equal contribution - A perceptually optimised bivariate visualisation scheme for high-dimensional fold-change data
Müller* A, Lausser* L, Wilhelm A, Ropinski T, Platzer M, Neumann** H, Kestler** HA
ADV DATA ANAL CLASSI 2021, https://doi.org/10.1007/s11634-0 * equal contribution, ** co-corresponding authors - Comprehensive Characterization of Multitissue Expression Landscape, Co-Expression Networks and Positive Selection in Pikeperch.
Nguinkal JA, Verleih M, de Los Ríos-Pérez L, Brunner RM, Sahm A, Bej S, Rebl A, Goldammer T
Cells 2021, 10(9), 2289 - Increased longevity due to sexual activity in mole-rats is associated with transcriptional changes in HPA stress axis.
Sahm* A, Platzer M, Koch P, Henning Y, Bens M, Groth M, Burda H, Begall S, Ting S, Goetz M, Van Daele P, Staniszewska M, Klose J, Costa PF, Hoffmann** S, Szafranski** K, Dammann** P
Elife 2021, 10, e57843 ** co-senior authors, * corresponding author