Subarea 5: Computational and Systems Biology of Aging
Subarea 5 focuses on the development of methods to analyse and understand complex biological systems. This work includes the design of computer algorithms and biostatistical approaches as well as the development of novel Omic strategies (i.e. genomics/epigenomics, transcriptomics, proteomics, and metabolomics) to study aging and aging-related diseases. According to the FLI, due to the Subarea's expertise in computational data analysis, it is deeply interconnected with all other Subareas. The Subarea hosts two critical core facilities (Life Science Computing, Proteomics) and provides consulting services in statistics. Furthermore, it organizes courses on data analysis and statistics.
The research is defined by five focus areas:
- Mapping extrinsic and intrinsic factors influencing stem cells during aging,
- Integration of spatiotemporal proteomics and transcriptomics data,
- Comprehensive evaluation of qualitative and quantitative expression changes,
- Identification and analysis of epigenomic alterations during aging and age-related diseases, and
- Network analysis of genomic, transcriptomic and epigenomic alterations during aging.
Research focus of Subarea 5.
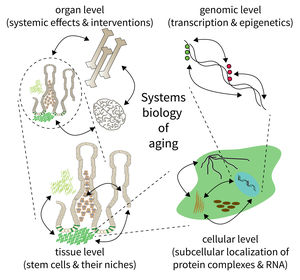
The biology of aging can be viewed as a multilayered array of networks at the level of organs, cells, molecules, and genes. The FLI wants to meet this complexity by establishing the new Subarea on “Computational and Systems Biology of Aging”. The overall goal is to interconnect research at different scales, taking place in Subareas 1-4 of the Institute’s research program. The new group on Systems Biology will integrate data from networks at multiple scales and will thus point to mechanisms and interactions that would not be seen in unilayer approaches.
Publications
(since 2016)
2020
- Constraining classifiers in molecular analysis: invariance and robustness.
Lausser L, Szekely R, Klimmek A, Schmid F, Kestler HA
J R Soc Interface 2020, 17(163), 20190612 - Detecting Ordinal Subcascades
Lausser* L, Schäfer* LM, Kühlwein SD, Kestler AMR, Kestler HA
Neural Process Lett 2020, 52, 2583–2605 * equal contribution - Chained correlations for feature selection
Lausser* L, Szekely* R, Kestler HA
ADV DATA ANAL CLASSI 2020, 14, 871–884 * equal contribution - Patterns of somatic structural variation in human cancer genomes.
Li Y, Roberts ND, Wala JA, Shapira O, Schumacher SE, Kumar K, Khurana E, Waszak S, Korbel JO, Haber JE, Imielinski M, PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Campbell PJ, PCAWG Consortium
Nature 2020, 578(7793), 112-21 - The GID ubiquitin ligase complex is a regulator of AMPK activity and organismal lifespan.
Liu H, Ding J, Köhnlein K, Urban N, Ori A, Villavicencio-Lorini P, Walentek P, Klotz LO, Hollemann T, Pfirrmann T
Autophagy 2020, 16(9), 1618-34 - ConCysFind: a pipeline tool to predict conserved amino acids of protein sequences across the plant kingdom
Moore M, Wesemann C, Gossmann N, Sahm A, Krüger J, Sczyrba A, Dietz KJ
BMC Bioinformatics 2020, 21(1), 490 - Aneuploidy-inducing gene knockdowns overlap with cancer mutations and identify Orp3 as a B-cell lymphoma suppressor.
Njeru* SN, Kraus* J, Meena* JK, Lechel A, Katz SF, Kumar M, Knippschild U, Azoitei A, Wezel F, Bolenz C, Leithäuser F, Gollowitzer A, Omrani O, Hoischen C, Koeberle A, Kestler** HA, Günes** C, Rudolph** KL
Oncogene 2020, 39(7), 1445-65 * equal contribution, ** co-corresponding authors - Genomic basis for RNA alterations in cancer.
PCAWG Transcriptome Core Group, Calabrese C, Davidson NR, Demircioğlu D, Fonseca NA, He Y, Kahles A, Lehmann KV, Liu F, Shiraishi Y, Soulette CM, Urban L, Greger L, Li S, Liu D, Perry MD, Xiang Q, Zhang F, Zhang J, Bailey P, Erkek S, Hoadley KA, Hou Y, Huska MR, Kilpinen H, Korbel JO, Marin MG, Markowski J, Nandi T, Pan-Hammarström Q, Pedamallu CS, Siebert R, Stark SG, Su H, Tan P, Waszak SM, Yung C, Zhu S, Awadalla P, Creighton CJ, Meyerson M, Ouellette BFF, Wu K, Yang H, PCAWG Transcriptome Working Group, Brazma A, Brooks AN, Göke J, Rätsch G, Schwarz RF, Stegle O, Zhang Z, PCAWG Consortium
Nature 2020, 578(7793), 129-36 - Vulnerability of progeroid smooth muscle cells to biomechanical forces is mediated by MMP13.
Pitrez PR, Estronca L, Monteiro LM, Colell G, Vazão H, Santinha D, Harhouri K, Thornton D, Navarro C, Egesipe AL, Carvalho T, Dos Santos RL, Lévy N, Smith JC, de Magalhães JP, Ori A, Bernardo A, De Sandre-Giovannoli A, Nissan X, Rosell A, Ferreira L
Nat Commun 2020, 11(1), 4110 - Analyses of non-coding somatic drivers in 2,658 cancer whole genomes.
Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, Hornshøj H, Hess JM, Juul RI, Lin Z, Feuerbach L, Sabarinathan R, Madsen T, Kim J, Mularoni L, Shuai S, Lanzós A, Herrmann C, Maruvka YE, Shen C, Amin SB, Bandopadhayay P, Bertl J, Boroevich KA, Busanovich J, Carlevaro-Fita J, Chakravarty D, Chan CWY, Craft D, Dhingra P, Diamanti K, Fonseca NA, Gonzalez-Perez A, Guo Q, Hamilton MP, Haradhvala NJ, Hong C, Isaev K, Johnson TA, Juul M, Kahles A, Kahraman A, Kim Y, Komorowski J, Kumar K, Kumar S, Lee D, Lehmann KV, Li Y, Liu EM, Lochovsky L, Park K, Pich O, Roberts ND, Saksena G, Schumacher SE, Sidiropoulos N, Sieverling L, Sinnott-Armstrong N, Stewart C, Tamborero D, Tubio JMC, Umer HM, Uusküla-Reimand L, Wadelius C, Wadi L, Yao X, Zhang CZ, Zhang J, Haber JE, Hobolth A, Imielinski M, Kellis M, Lawrence MS, von Mering C, Nakagawa H, Raphael BJ, Rubin MA, Sander C, Stein LD, Stuart JM, Tsunoda T, Wheeler DA, Johnson R, Reimand J, Gerstein M, Khurana E, Campbell PJ, López-Bigas N, PCAWG Drivers and Functional Interpretation Working Group, PCAWG Structural Variation Working Group, Weischenfeldt J, Beroukhim R, Martincorena I, Pedersen JS, Getz G, PCAWG Consortium
Nature 2020, 578(7793), 102-11